Targeted LC-MS/MS
Table of contents
Background
- Applications of targeted LC-MS analysis are diverse and include disease biomarker discovery, pharmaceutical research, nutritional studies, and personalized medicine. It contributes to a better understanding of metabolic dysregulation in various health conditions and plays a pivotal role in advancing our knowledge of the metabolome.
- The selection of specific analytes is based on prior knowledge or hypothesis about the compounds of interest.
- These analytes could be drugs, metabolites, proteins, peptides, environmental contaminants, or any other molecules of interest.
- The goal is to accurately determine the presence and concentration of these selected analytes with a high level of sensitivity and specificity.
- The workflow of targeted LC-MS analysis in metabolomics involves sample preparation, chromatographic separation, mass spectrometric detection, and data analysis.
- Stable isotope-labeled internal standards and calibration curves are employed to improve the accuracy of metabolite quantification.
- In conclusion, targeted LC-MS analysis is a specialized and powerful approach that enables to investigate specific metabolites within complex biological systems. It has become an indispensable tool for gaining deeper insights into metabolic processes and their implications in health and disease.
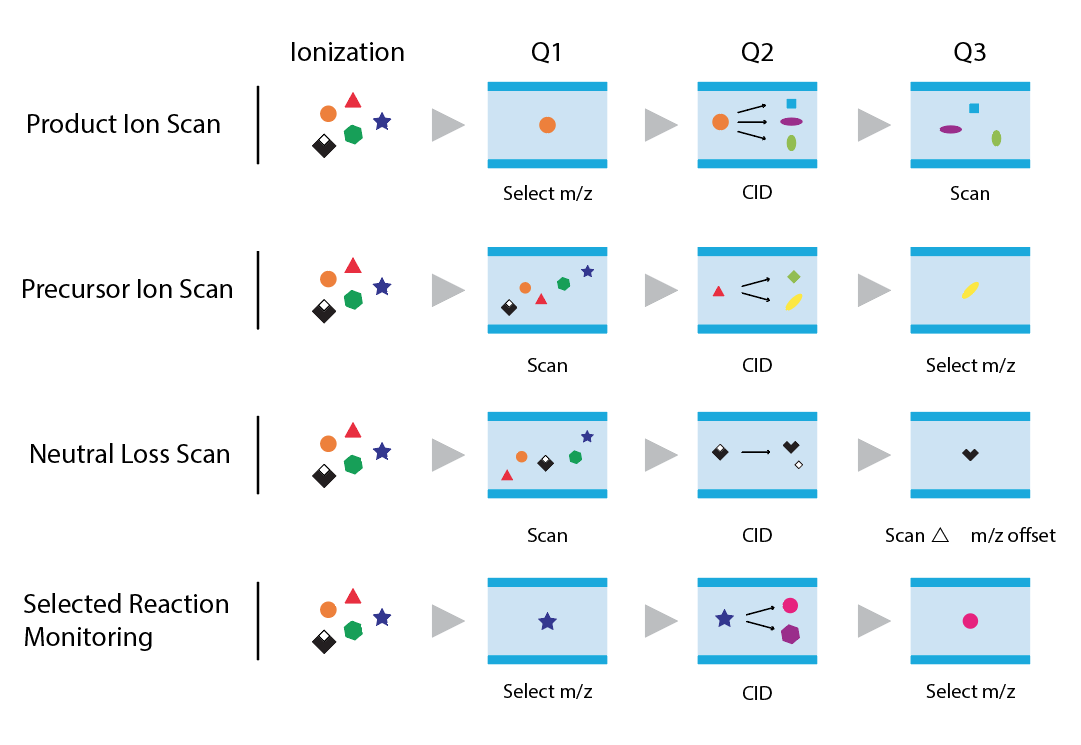
SRM method development
- Selected Reaction Monitoring (SRM) is a widely used mass spectrometry technique for targeted quantification of specific analytes in complex samples. During method development, metabolite standards are fragmented at different energies to create a catalog of all observable fragments. This information is used to identify fragments, which allow discriminating the compound from others with similar precursor mass. Data for this scan mode can also be retrieved from public repositories e.g (Metlin or MzCloud. A targeted run can include several hundreds of such SRM scans in a multiple reaction monitoring (MRM) method. Here is an overview of SRM method development:
- Analyte Selection: The first step in SRM method development is the selection of target analytes (e.g., peptides or small molecules) that are of interest for quantification. The analytes are chosen based on the individual research question.
- Precursor-Product Ion Pairs: For each selected analyte, precursor and product ion pairs are determined. The precursor ion is the intact molecular ion of the analyte, and the product ions are the fragments generated during collision-induced dissociation (CID) in the mass spectrometer. These pairs are specific to each analyte and serve as the basis for quantification.
- Optimization of Parameters: Key instrument parameters, such as collision energy, collision cell gas pressure, polarity, spray voltage and dwell time, are optimized to maximize the sensitivity and selectivity for each precursor-product ion pair. This involves infusing the pure standard of each compound to find the ideal settings.
- Method Validation: The SRM method is validated by assessing its linearity, sensitivity, accuracy, precision, and selectivity. Calibration curves and quality control samples are often used for validation.
- Acquisition and Data Processing: The mass spectrometer is programmed to perform SRM acquisitions, cycling through the precursor-product ion pairs for all the target analytes. The acquired data is processed using dedicated software, which quantifies the analytes based on their ion intensity and the calibration curves. See: No such attachment on this page.
PRM method development
- Parallel Reaction Monitoring (PRM) is a powerful and advanced mass spectrometry-based technique used for targeted analysis in metabolomics. PRM allows for the simultaneous monitoring and quantification of multiple specific compounds in a complex sample. Its development involves several essential steps:
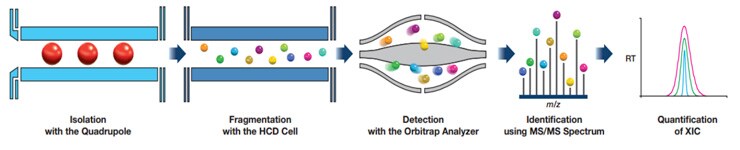
- PRM Transition List: A transition list is generated, which specifies the precursor ions and their corresponding product ions (transitions) for each selected molecule. These transitions are critical for mass spectrometric detection and quantification.
- Instrument Optimization: The mass spectrometer is optimized for PRM analysis. Parameters, such as collision energy and isolation width, are tuned to maximize sensitivity and selectivity for each transition.
- Method Validation: The SRM method is validated by assessing its linearity, sensitivity, accuracy, precision, and selectivity. Calibration curves and quality control samples are often used for validation.
- Acquisition Method: The acquisition method is set up to specifically target the precursor-product ion transitions from the transition list. In PRM, all transitions are monitored simultaneously, allowing for high sensitivity and selectivity.
Data Analysis
- Acquired raw data files are processed, analyzed and quantified with the application Quan Browser in the Thermo Scientific Xcalibur software suite. A detailed description of the whole process with an exhaustive coverage: No such attachment on this page.
Method Validation Concepts
- Method validation in targeted LC-MS is a critical aspect and encompasses a series of processes and criteria to confirm the fitness of the analytical method for its intended purpose. This includes evaluating parameters like selectivity, sensitivity, accuracy, precision, linearity, and robustness. The validation process ensures that the method can consistently and accurately measure the desired analytes, even in complex matrices. It is good practice to follow the recommendations of the Food and Drug Administration: No such attachment on this page.